Leonard M. Adleman, the A.M. Turing Award Laureate and one of the creators of the RSA encryption, is widely referred to as the Father of DNA Computing. The field of DNA computing is generally considered to have begun with Leonard Adleman’s 1994 experiment involving the “Hamiltonian Path problem”, but is more popularly recognized as a variant of the so-called “traveling salesman problem(or “TSP” for short).”
There is no better way to understand how DNA computing works than by going through an example step by step. So let’s solve our own directed Hamiltonian Path problem, using the DNA methods demonstrated by Adleman. The concepts are the same but the example has been simplified by me to make it easier to follow and present.
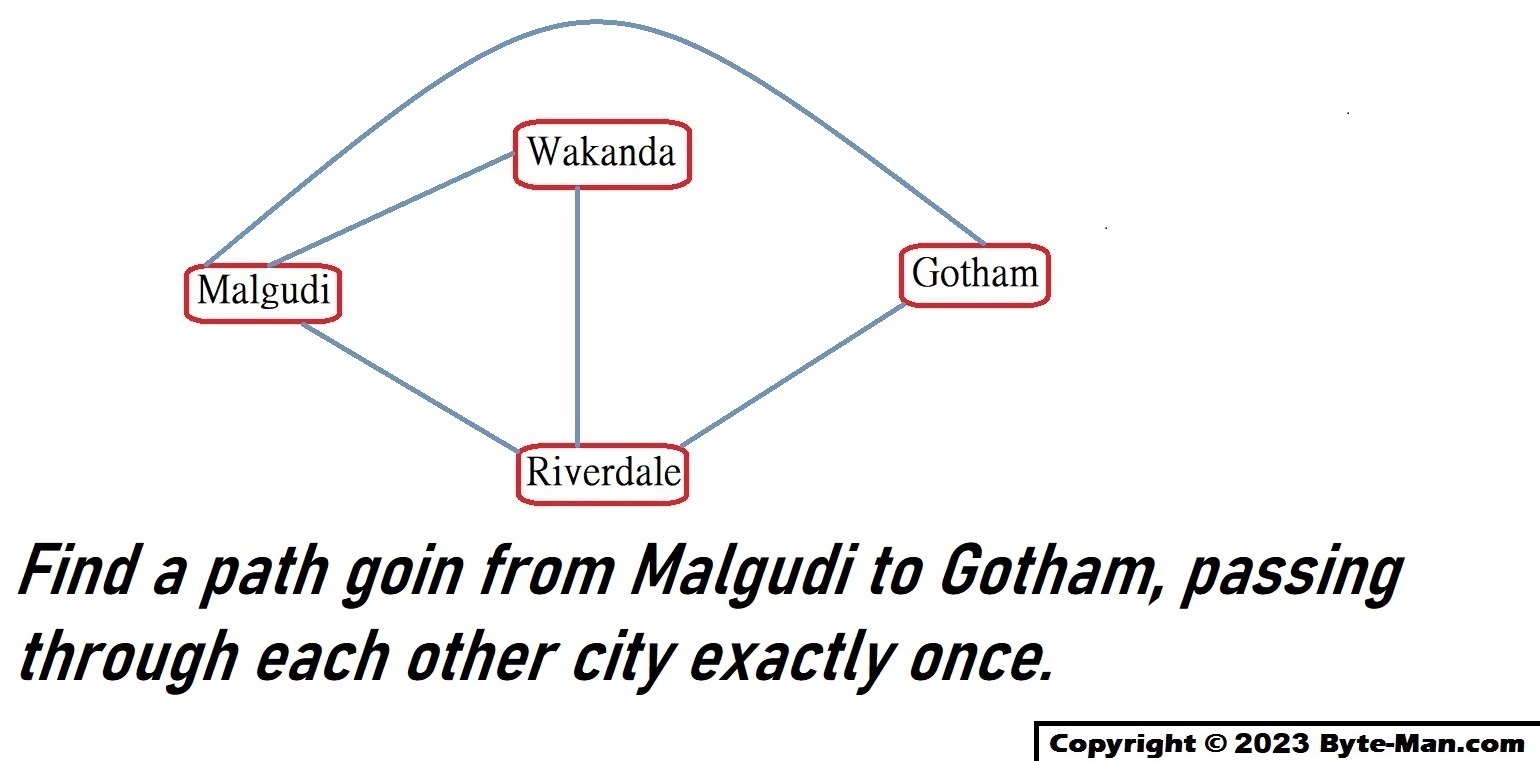
We are going to solve traveling salesman problem involved four cities using DNA computing. Find a path going from Malgudi to Gotham, passing through each other city exactly once.
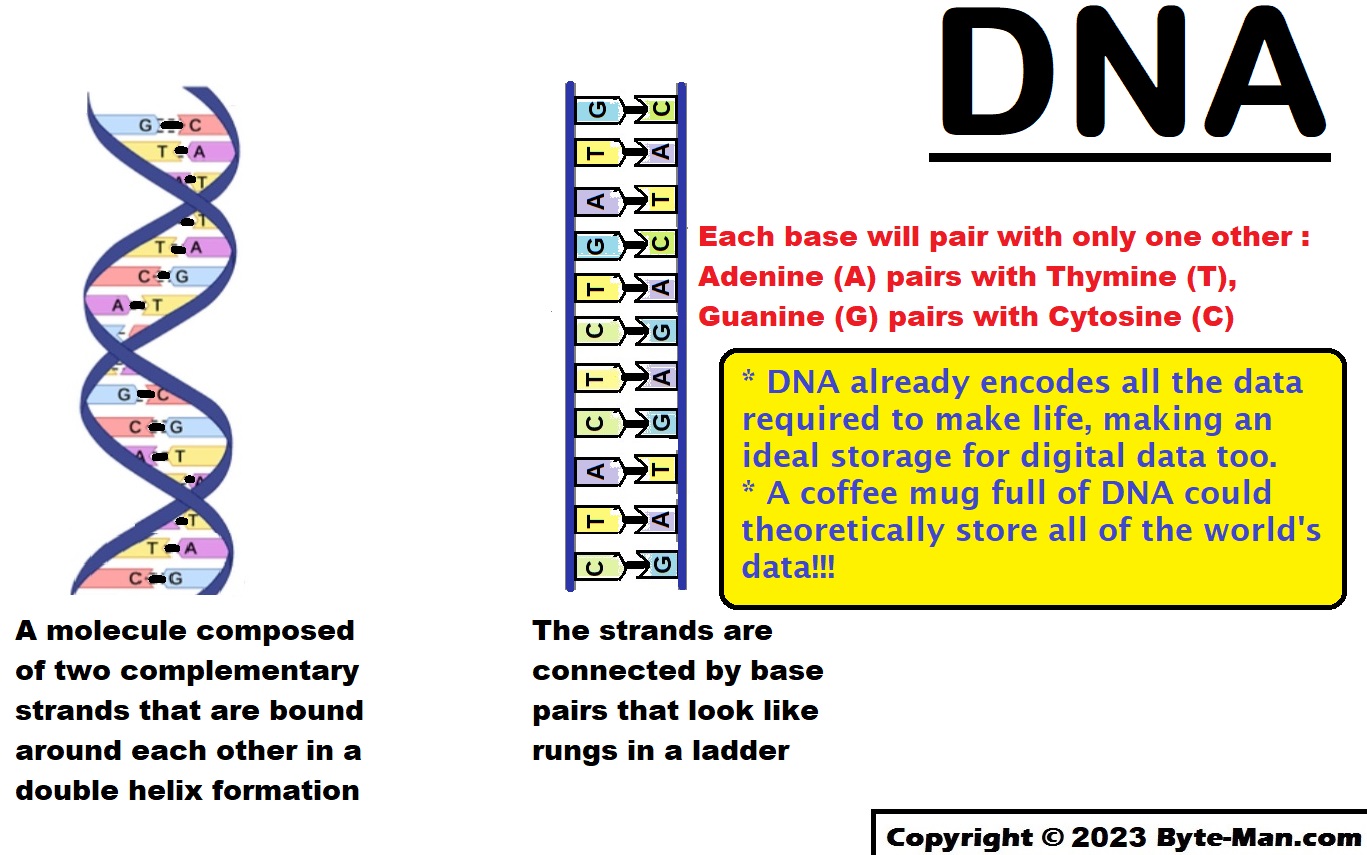
DNA stands for deoxyribonucleic acid. Each organism’s DNA is unique and it basically contains the blueprint for every living organism, determining how each organism looks and functions. In simple words, our data are stored in DNA. DNA is an attractive material for data storage – it is stable, writable, readable, and information dense.
The information in DNA is stored as a code made up of four chemical bases: adenine (A), guanine (G), cytosine (C), and thymine (T). DNA bases pair up with each other, A with T and C with G, to form units called base pairs.
Solving Hamiltonian Path Problem Using DNA Computing
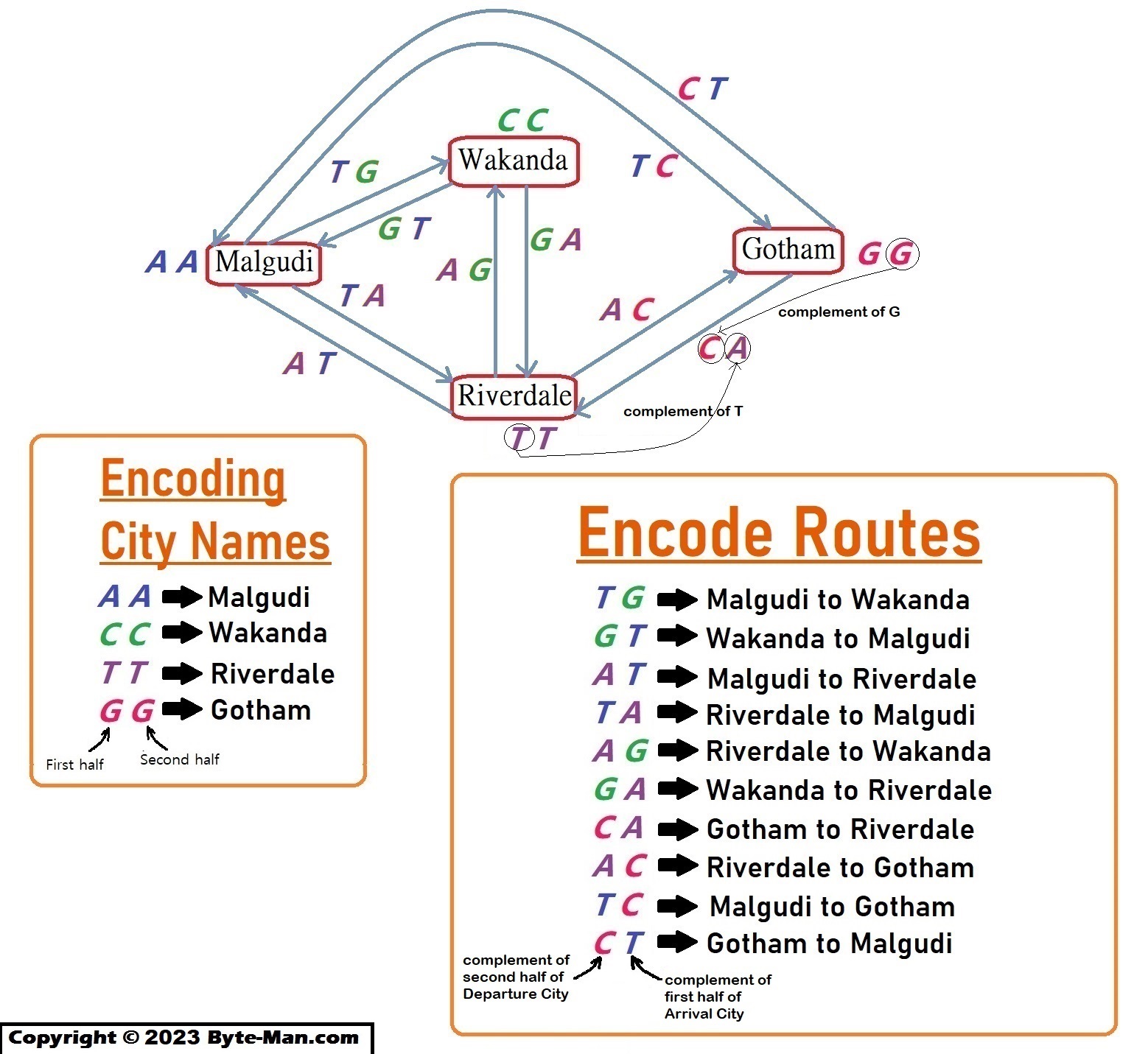
Part I: Encode city names in short DNA sequences.
DNA can simply be treated as a string of data. For example, each city can be represented by a letter or a word of DNA bases.
Part II: Encode the routes between the cities by a DNA fragment.
A route between two cities can be encoded by the complement of second half(last letter) of the departure city and the first half(first letter) of the arrival city.
Part III: Generate all possible routes.
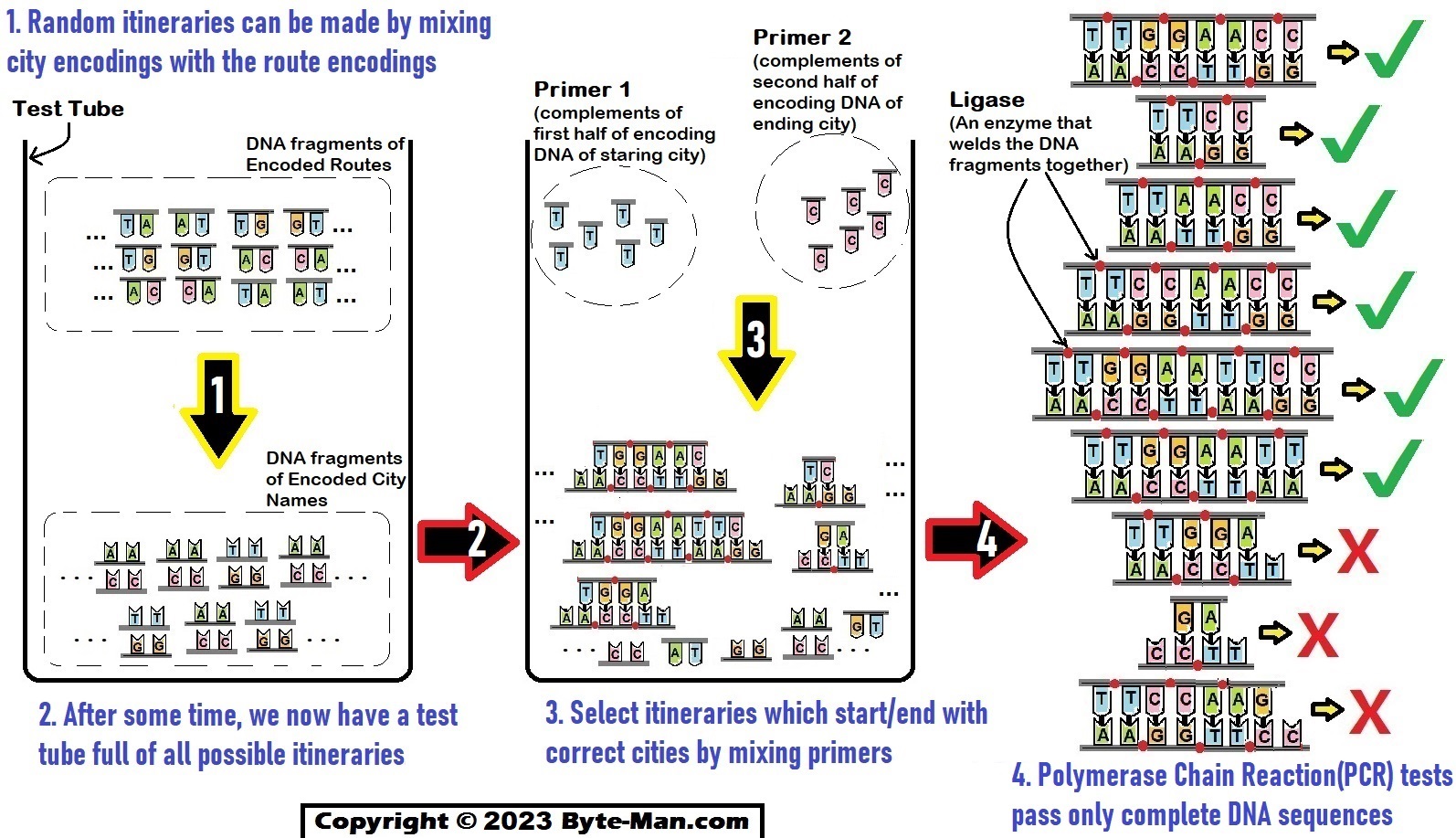
Random itineraries can be made by mixing city encodings with route encodings in a test tube.
Part IV: Select itineraries which start/end with correct cities
The diagram shows how selective PCR(polymerase chain reaction) pass fragments with the proper start/ending points from all DNA strands.
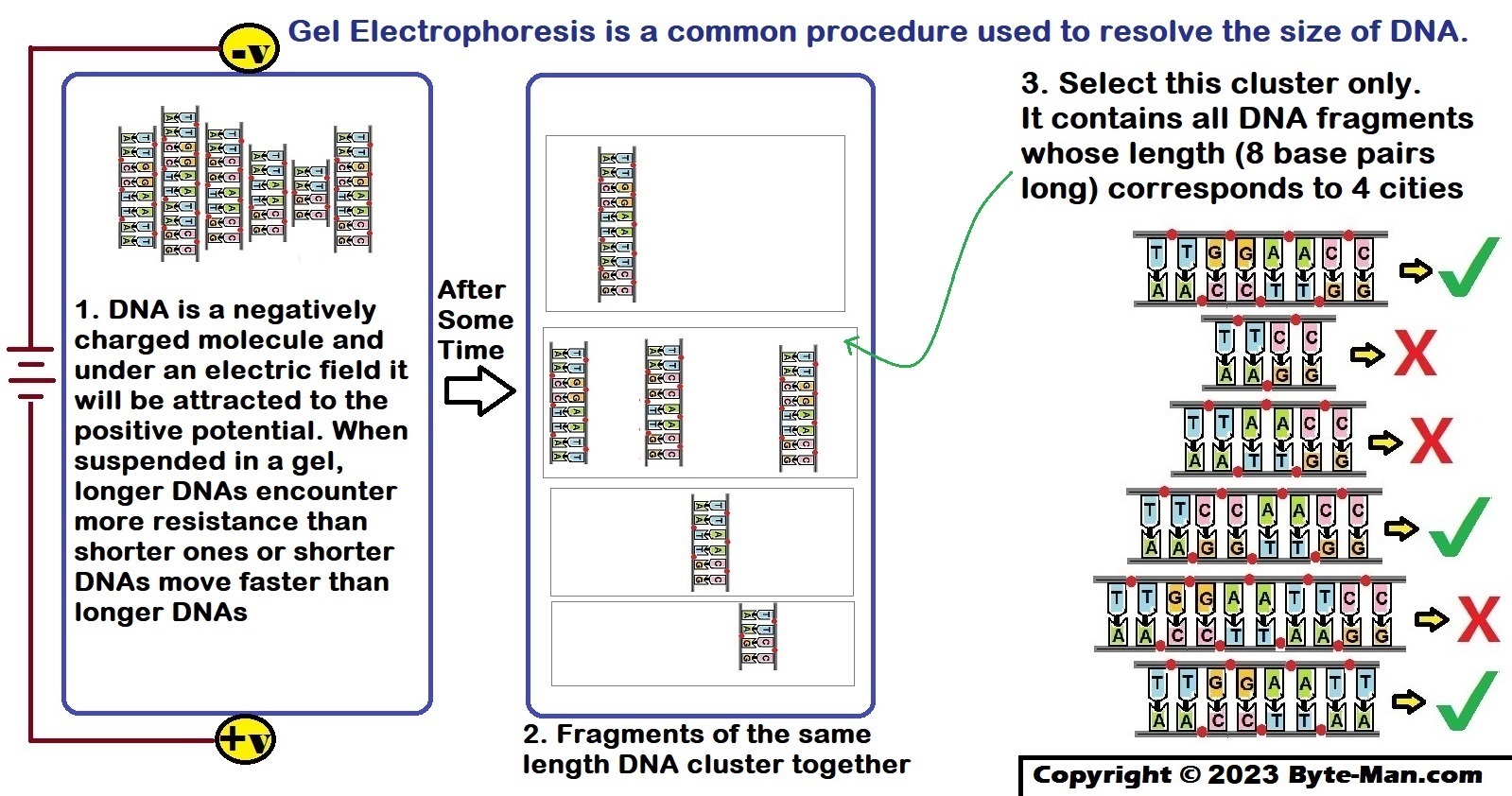
Part V: Select itineraries that contain the correct number of cities
Sort DNA by length and select the DNA whose length corresponds to 4 cities.
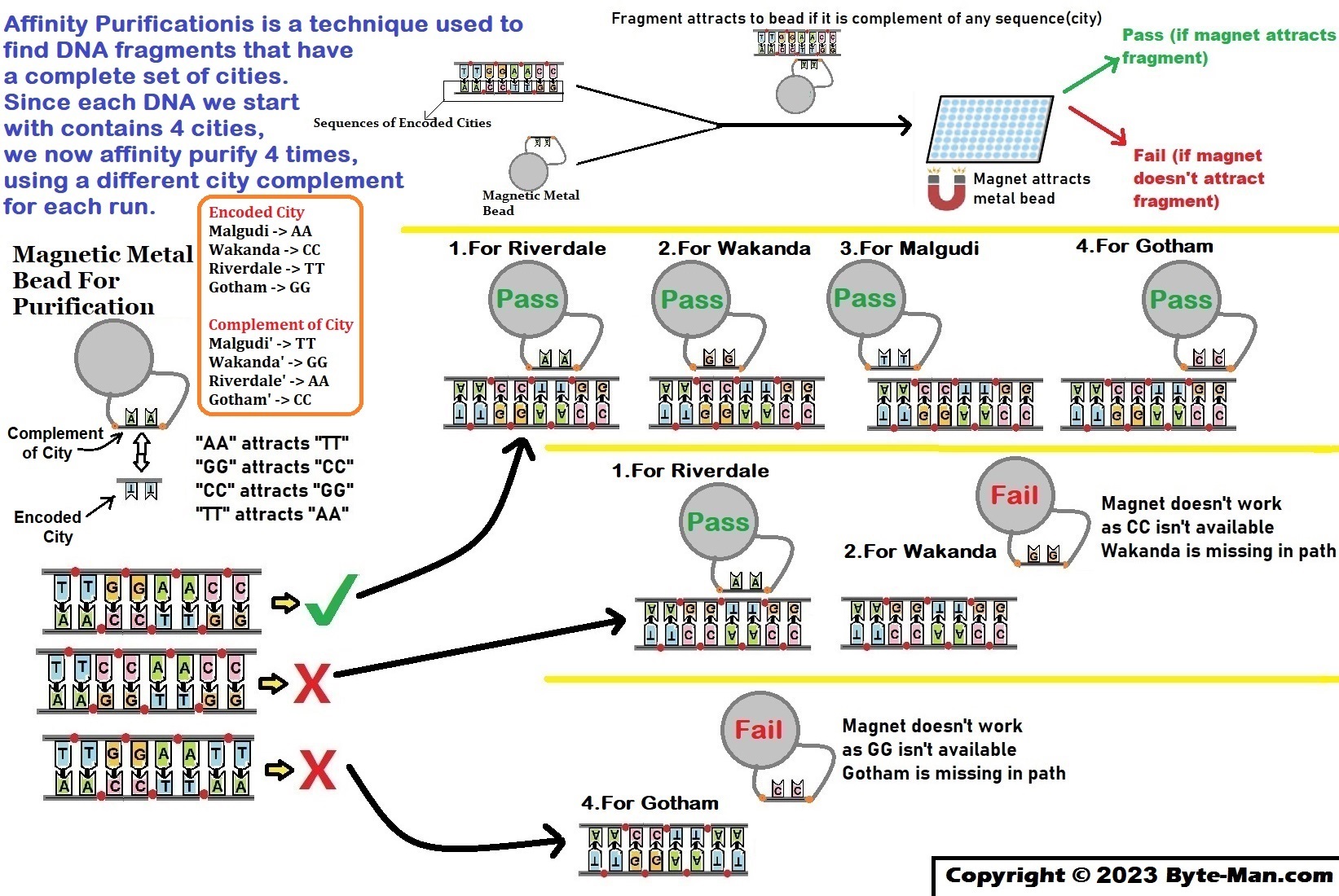
Part VI: Select itineraries that have a complete set of cities
Select the DNA which contains all 4 cities – Malgudi, Wakanda, Riverdale & Gotham.
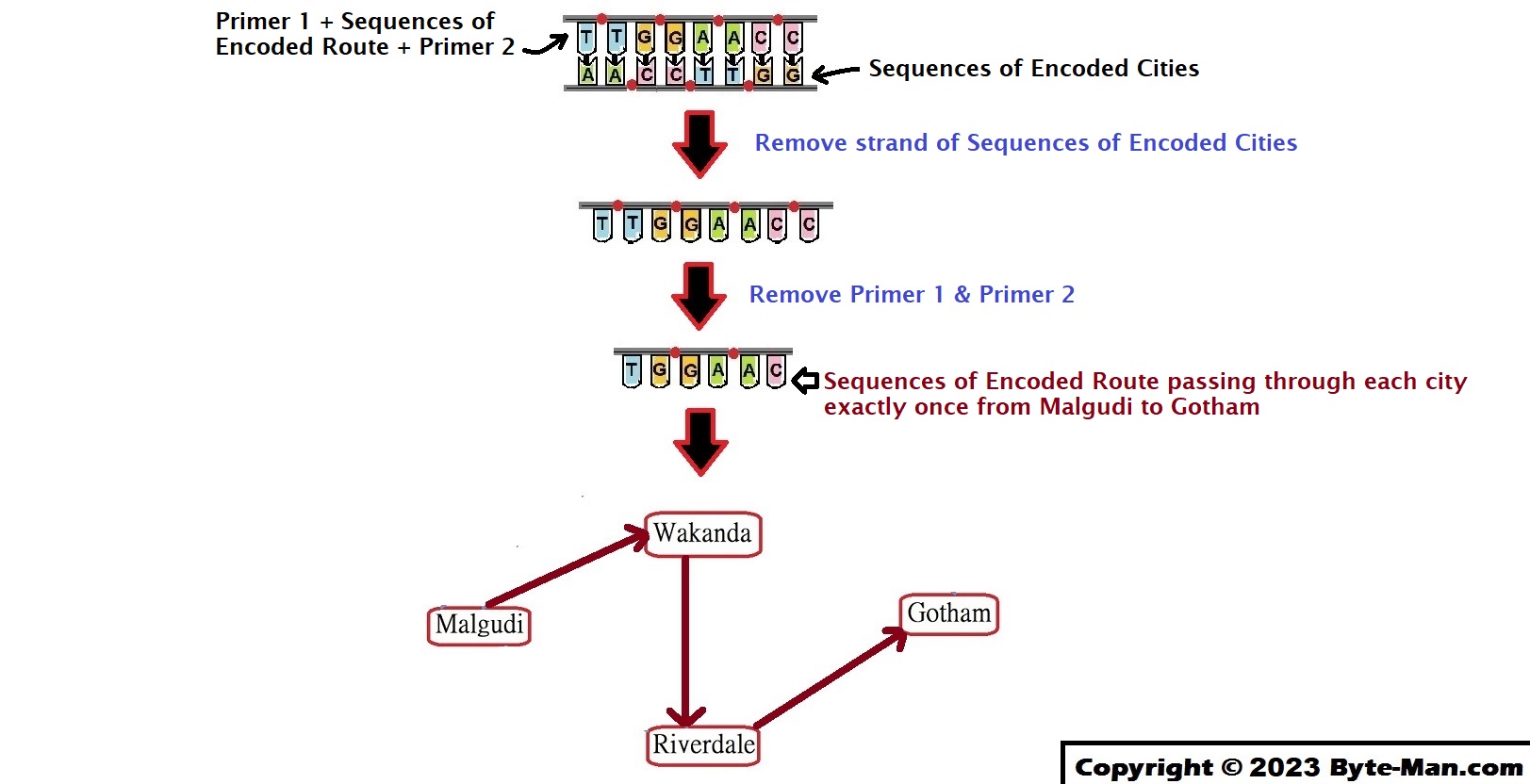
Each fragment that passed the PCR test starts/ends at the right place;
Each fragment that passed the gel electrophoresis test has the right number of cities;
Each fragment that passed affinity purification test has a complete set of cities.
So, the remaining DNA fragment(s) which passed all the tests must be our solution, Hamiltonian!!!
In reality Adleman’s experiment was much more complex and included 7 cities. As the number of cities increases, the problem becomes more difficult and expensive to compute. TSPs with a large number of cities are impractical to solve on even the latest supercomputer. Adleman’s experiment with DNA computing demonstrated unique aspects of DNA as a data structure and showed that one of the hardest problems a computer can encounter has been solved by using DNA molecules.
PS : Graphical representation was added in this article on February 2023.